Sequence Database Setup: MSDB
Overview
The MSDB database is compiled by the
Proteomics Group at Imperial College London.
MSDB is a composite, non-identical protein sequence database built from a number of primary source databases.
Sequences from the higher priority databases are preferentially retained. The present source databases
(in order of priority) are:
PIR,
Trembl,
GenBank,
Swiss-Prot, and
NRL3D.
One of the main advantages of MSDB is the accompanying full-text reference file.
Download
ftp://ftp.ebi.ac.uk/pub/databases/MassSpecDB/
ftp://ftp.ncbi.nih.gov/repository/MSDB/
ftp://csc-fserve.hh.med.ic.ac.uk/pub/
You should download three files: the Fasta database (msdb.fasta.Z), release notes (msdb.nam.Z),
and a reference file. There is a choice of reference files: a complete reference file including Swiss-Prot
annotation text, (msdb.ref.complete.Z), and a file in which the Swiss-Prot annotations have been
replaced by links to the Expasy web site, (msdb.ref.Z). This dates from when a licence was required for
commercial use of Swiss-Prot.
To download updates automatically, the relevant definition block in
db_update.pl is either MSDB_from_EBI or MSDB_from_NCBI.
Taxonomy
Taxonomy for MSDB is predefined in mascot.dat, choose "MSDB REF".
The following taxonomy files are required:
ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
Note that the taxonomy files go into the taxonomy directory, not into the sequence database
directory. Also, some files need to be unpacked (using tar) as well as uncompressed.
Parse Rules
A typical Fasta title line is:
>B32382 fcbH bifunctional protein -
Bradyrhizobium japonicum
The single identifier varies according to the source database. Suitable parse rules are:
Accession from Fasta title: ">\([^ ]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
The corresponding line in the Ref file is:
>P1;B32382
Accession from Ref file: ">[A-Z][0-9];\([^ ]*\)"
Configuration
For this example, the three files were downloaded to a folder named
C:\Inetpub\MASCOT\sequence\MSDB\current.
The files were decompressed using gzip,
and renamed to MSDB_20020515.fasta, ref, and nam.
When updating an active database, it is important to rename the Fasta file last, because Mascot
will begin database exchange as soon as it sees a new Fasta file that matches the wildcard path for
the database.
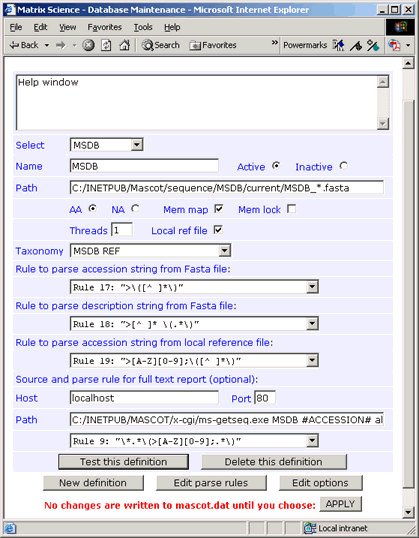
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port, and Path fields blank
and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|